The R-Extension of NetLogo provides primitives to use the statistical software R (Gnu S) (see the R Project website) within a NetLogo model. There are primitives to create R-Variables with values from NetLogo variables or agents and others to evaluate commands in R with and without return values.
To use the extension in your model, add a line to the top of your procedures tab:
extensions [ r ]
If your model already uses other extensions, then it already has an
extensions line in it, so just add r to the list.
For more information on using NetLogo extensions, see the Extensions Guide.
For examples of the usage of the R-Extension, models can be downloaded from the project repository. These models are installed with NetLogo in the “models” directory of the R extension. Please note that (as of NetLogo 6.0) these models are not included in the NetLogo models library.
If you want to use the plot function of R, you could activate the JavaGD plot device via r:setPlotDevice, see the “plot-example1.nlogo” model. This is the prefered method!
But you can also use the standard R device, but then, you have to give R some cpu time, e.g.
by run an evalulation of sys.sleep(0.01) with a forever button. See the
“plot-example2.nlogo”. (Many thanks to Thomas Petzold!). The creation of plots into
files is also possible. See the “plot-into-file-example.nlogo” in the examples folder.
It’s possible to load and save data from file directly in R. This code snippet illustrates:
r:eval "dataname <- read.table('<path to file>')" ; read file
r:eval "write.table(dataname, file='<filename>')" ; write file
Normally, a data.frame cell contains only a single value. Each column is represented as a
vector and if you would put a vector of vectors to a data.frame, it would be splitted into
several columns. With the R-Extension it is possible to put a vector into a data.frame cell,
when you assign a NetLogo List to a column which contains nested NetLogo Lists for each row.
If you want, for example, to use write.table on this data.frame, you have to mark this
column as class="AsIs". You can do this by using the I(x)-function.
Example: If the column of interest has the name “col1” of the data.frame “df1” you could
execute r:eval "df1$col1 <- I(df1$col1)". Call help(I) from within an R
terminal for further details.
Furthermore, you can define functions in an R-Script, load it, and use the functions.
Load R-files via r:eval "source('<path to r-file>')".
It’s also possible to load R packages via r:eval "library(<name of package>)".
When you compile your code containing extensions [r] you will create a new R
workspace. Until you reload the extension, open a new model or submit the primitive
[r:clear](#rclear), all R variables assigned in this session will be available like you would
use R from the command line or in the R Console.
You can open an Interactive R Shell via r:interactiveShell. This shell is a port to
the underlaying R instance. This shell works on the global environment (see Environments in the R Extension below) while the extension itself work on a custom local
environment. But there is one automatic variable “nl.env” in the global environment, which
is a reference to the local environment of the extension. Don’t delete this variable!
You can access a variable created by the extension via
get("<variable name>",nl.env), for example myvar <- get("myvar",nl.env).
If you want to plot from the Interactive Shell you should use the included JavaGD plot
device (see r:setPlotDevice).
You can save and load the history of entered R commands via a right-mouse button context
menu.
Please read the notes at the top of the output text area after opening the shell! On Linux OS it can happen that you see an error message from X11. Please check, if everything worked correcly. If so, you can ignore these messages. If not, please write a report to bugs@ccl.northwestern.edu or open an issue.
When you load a model the R-Extension creates a new R environment. When you create an R
variable using the R-Extension, this variable is created in the local R environment.
Furthermore, all calls from the R-Extension work on this local environment. This new
environment concept enables you to use the extension in BehaviorSpace Experiments.
Therefore, you don’t have to care about the environment while you’re not using the
Interactive Shell or other tools, which work on the global environment. You can explicitly
assign a variable to the global environment by using the <- operator or by executing
assign(<name>,<value>,envir=.GlobalEnv). If you work with the Interactive Shell,
see the notes at the top of the output text area after opening the shell.
Type help(environment) in an R shell to learn more about environments.
You can/should clear (i.e. remove all variable and free memory) the local environment via
r:clearLocal. If you want to clear also the global environment (the whole workspace), call
r:clear.
With the R-Extension you can load R into the process of NetLogo. Because of the architecture of R, both software share one system process and therefore the memory given to NetLogo.
In some circumstances it can happen that you receive an out of memory error due to Java’s heap space. You can increase the heap space before starting NetLogo by adapting the -Xmx JVM-parameter (see also the NetLogo manual section on Windows memory). But on 32-bit systems, this is very limited. Therefore, it is a good idea to use a 64-bit system if you want/need to use high amount of RAM.
You can see the memory usage of R by starting the interactive shell (r:interactiveShell) and type there:
memory.size(max=F) and memory.size(max=T). Furthermore, you can check the memory limit by typing: memory.limit().
See also:
If you call the garbage collector in the interactive shell by typing gc(), you will get some information about the current memory usage (see also http://stat.ethz.ch/R-manual/R-patched/library/base/html/gc.html).
If you type gc(nl.env) you will see the percentage of memory used for cons cells and vectors.
Don’t forget to call the r:gc primitive after removing an R variable and don’t forget to remove R variable you don’t need anymore! See how the memory usage changes after removing variable and calling r:gc.
If you use too much memory, it can happen, that NetLogo will close abruptly. In such a case, check if there is a way to reduce the memory used. If not, try to switch over to the Rserve-extension. With the Rserve-Extension both software, NetLogo and R, run independently. There is, of cause, also a limit of transferable data amount with one request, but it is less restrictive.
One last note to this topic: Keep in mind that R is a vector-oriented language. Prevent mass calls with single values whenever possible and replace them by vector operations. This is much faster and more stable.
Since R-Extension version 1.1 it is possible use the extension when NetLogo is running in headless mode.
This is for example the case, when you run BehaviorSpace experiments from the command line (see here).
The difference is, that the interactiveShell is not initialized/instanciated.
You can use the extension as you know it from GUI mode, but it is not possible to open the interactiveShell (r:interactiveShell) and to set the plot device (r:setPlotDevice).
But one additional things has to be done: You have to call r:stop finally when running NetLogo headless to stop the R engine. Otherwise NetLogo will not be closed and you will not get back to the command line prompt.
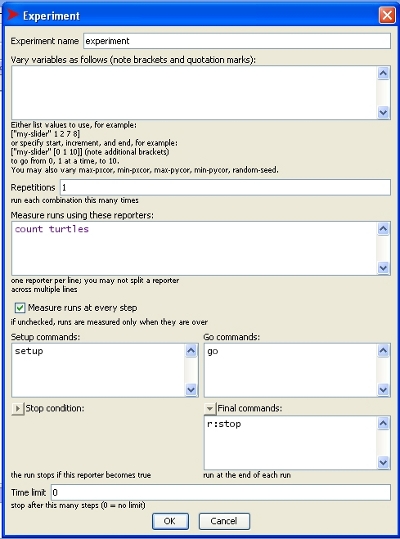
When setting up a BehaviorSpace experiment, there is the option to set final commands.
This is a good place to add the r:stop command (see image).

The R Extension is bundled with NetLogo 6. To use it, you will need a compatible R installation and you may need to configure the extension.
Standard R 3 installations should work (sometimes without configuration). As of NetLogo 6.0.2, the following operating system / R versions were tested:
Once R is installed, you will need to install the rJava package.
Certain features of the R extension rely on the JavaGD package.
To install, start the RGui from your program list, click on the item “Packages” in the menu bar and then on “Install Package(s)”. Select your favorite server and find “rJava”, as well as “JavaGD” and/or “CommonJavaJars” (both optional) in the list of packages.
If you prefer using the console, you can install the same packages by running the following commands in the console (and following the prompts they generate, as appropriate).
install.packages("rJava")
install.packages("JavaGD") # Optional
install.packages("CommonJavaJars") # Optional
If you are using Linux or Mac OS and one of the above R versions, you may not need to
perform any further configuration.
An easy way to determine whether you need to configure the extension it to open a new NetLogo model,
add extensions [ r ] to the code tab and press “Check.”
If you see an error, you need to configure the R extension.
The R extension can be configured by editing the “user.properties” file in a text editor.
You should place the “user.properties” file in your user folder as indicated by the error message you get when R is not configured.
The per-system user folders are typically:
/Users/YOUR_USERNAME/Library/Application Support/NetLogo/6.1/r/C:\Users\YOUR_USERNAME\AppData\Roaming\NetLogo\6.1\r\/home/YOUR_USERNAME/.netlogo/6.1/r/The following keys are used to configure the extension:
r.home: Controls which installation of r is used.jri.home.paths: Controls the path to the jri subdirectory of the rJava library.Note that you will have to exit NetLogo and restart to see configuration changes take effect,
as the configuration file is only loaded once per NetLogo instance.
See below on how to determine the appropriate values to for r.home and jri.home.paths.
r.home and jri.home.pathsr.home is the path to the “R” installation directory which contains the “bin” directory.
If you’re having trouble finding this, you can run R.home(component = "home") in R, or
R RHOME on the command line (if R is on your path).
R.home(component = "home")
# Returns "C:/PROGRA~1/R/R-36~1.1" on Windows.
# Will return other results on other platforms or configurations
jri.home.paths is a list of directories to check for jri.
It’s in the jri directory under the rJava library installation.
You can find the jri directory in the rJava package by running the following in R:
system.file("jri", package = "rJava")
# Returns "C:/Users/USER_NAME/Documents/R/win-library/3.6/rJava/jri" on Windows.
# Will return other results on other platforms or configurations
Take the path and edit the user.properties file, uncommenting and editing one set of r.home and jri.home.paths to match
the values obtained in R.
When you’re done, the user.properties file should have the following lines (given the above results):
r.home=C:/PROGRA~1/R/R-36~1.1
jri.home.paths=C:/Users/USER_NAME/Documents/R/win-library/3.6/rJava/jri
Save user.properties and load a model using the R extension. You should see it start and run properly.
The R Extension on Windows requires the Microsoft Visual C++ 2013 redistributable to be installed. You can get that as a free download from Microsoft.
Windows also requires the additional configuration step of configuring the PATH environment variable. Additionally, editing the user.properties file on Windows is slightly more difficult than on other platforms.
To begin, determine the appropriate directory from your R installation to add to your PATH.
To do this, determine where your R installation is located (here we’ll use the location C:\Program Files\R\R-
C:\Program Files\R\R-<version>\bin\i386\C:\Program Files\R\R-<version>\bin\x64\Note that you will need to update this setting if you wish to upgrade the version of R used by NetLogo.
“user.properties” is a newline-delimited file. This means if it is opened in “Notepad” it will look like all the text is on a single line. For this reason, it is recommended to open first in “WordPad” and resave before editing in Notepad. Alternatively, if you have a full-featured text editor (like Notepad++, Vim, or Emacs) installed, you can use that to edit the file.
To reiterate a warning given in the “user.properties” file, the directory separator for Windows must be entered in user.properties as double-backslash (“\”) or single-forward-slash (“/”).
r:clear
r:clearLocal
r:eval
r:__evaldirect
r:gc
r:get
r:interactiveShell
r:put
r:putagent
r:putagentdf
r:putdataframe
r:putlist
r:putnamedlist
r:setPlotDevice
r:stop
Clears the R-Workspace. All variables in R will be deleted. It evaluates the R command
rm(list=ls()) and rm(list=ls(nl.env)).
This deletes variables created in global as well as local environment
(see R Environments for details about environments).
It’s always a good idea to add this command to your setup procedure under your “clear-all” call.
;; clear the R workspace
r:clear
It clears the local R environment, which is used by the extension. All variables which have
been created in the local environment will be deleted. It evaluates the R command
rm(list=ls(nl.env)).
See R Environments for details about environments.
See r:clear for deleting all variables, i.e. the globals as well.
;; delete the local variables
r:clearLocal
It evaluates the submitted R command. The R command shouldn’t return a value.
;; creates a new vector in R with a sequence from 1 to 10
r:eval "x <- seq(1,10)"
show r:get "x"
Evaluates the submitted R command in the global environment (not in the local environment like r:eval does) and
without a check (not using try-function internally).
This can be necessary for some R packages, like gglopt2.
Please note, that you can produce name clashes when creating new variables using this primitive.
The variable will be created into the global environment and will not overwrite variable with the same name that have been created into the local environment.
If you request a variable with r:get it will search in the local environment first.
Therefore, if there are variables with the same name in the local and the global environment, it will report the variable from the local environment and not the variable created via r:__evaldirect.
If there is only a variable with the requested name in the global environment, everything will be fine - r:get will report the value of this variable.
If you want to remove a variable created via r:__evaldirect, i.e. in the global environment, call r:eval "rm(myvar, envir=.GlobalEnv)", replace myvar by the name of your variable.
The R command shouldn’t return a value.
This primitive is experimental.
;; creates a new vector in R with a sequence from 1 to 10
r:__evaldirect "x <- seq(1,10)"
show r:get "x"
Calls the garbage collector of Java (i.e. the R-Extension) and R. Call this primitive after removing an R variable to free the memory.
;; create a variable
r:eval "x <- 1:10"
;; remove the variable
r:eval "rm(x)"
;; call the garbage collector
r:gc
Reports the return value of the submitted R command. Return type could be a String, Number, Boolean, NetLogo List or a NetLogo List of Lists.
R lists will be converted into a NetLogo List. If the R list itself contains further lists, it will be converted into a NetLogo List with nested NetLogo lists. Lists containing values of different data types are also supported (e.g. mixed Strings, Numbers and Booleans/Logicals).
Data.frames will be converted into a NetLogo List with nested List for each column, but the column names will be lost (same for named R lists).
R matrices can be received, but they are converted into one NetLogo list. NULL and NA values are not converted and will throw an error, because NetLogo has no corresponding value.
;; returns a list with 10 variables
show r:get "rnorm(10)"
Opens a window with two textareas. The upper one is the R output stream and in the lower one you can type R commands. This is the access to the underlaying R session. You can type multi-line commands. To submit commands press Ctrl+Enter. With “PageUp” and “PageDown” in the input area you can browse through the histroy of submitted commands. With right-mouseclick context menu, you can save and load an RHistory (interchangeable with R terminal and other R GUIs).
Please note, that the Interactive Shell works on the global environment, while commands
submitted from NetLogo lives in an local environment. A reference to this local environment
is automatically added to the global environment (named nl.env, please do not delete this
variable. With a call of r:clear you can restore it but this will
empty your workspace). You can use this to have access to variables which you have created
from NetLogo by get("<variable name>",nl.env). To copy for example an variable
with the name var1 from the local environment to the global environment, type var <- get("var",nl.env). See section R Environments for details.
If you just want to see the contents of a variable which lives in the local environment, you
could submit your command, for example in the NetLogo Command Center, and the result will
be shown in the output area of the Interactive Shell. For example:
r:put "test" (list world-width world-height)
r:interactiveShell
r:eval "print(test)"
r:eval "str(test)"
Variables which have been created in the Interactive Shell are available from NetLogo, even if they are created in the global environment. But if there is a variable with the same name in the local environment, you will get this variable in NetLogo instead the one from the global environment.
If you want to execute plot commands from the Interactive Shell you should activate the
integrated JavaGD plot device via r:setPlotDevice first.
;; opens Interactive Shell
r:interactiveShell
Creates a new variable in R with the name name. The value can be a String, Number, Boolean or List.
NetLogo Lists are converted to R vectors, if all entries are of the same data type. If a NetLogo list contains different data types (mixed Strings, Numbers of Booleans), it will be converted into an R list. If a NetLogo List contains other/nested NetLogo Lists it will be converted into an R list and the nested Lists are handled by the same rule (Vectors if all items are of the same data type, …).
;; creates an R variable "testvar" with the size of turtle 0
r:put "testvar" [size] of turtle 0
show r:get "testvar"
Creates a new named list in R with the name name.
The argument variables is any number of strings which list and variable(s) of the agent|agentset.
Names of the elements of the R list will be the same as the names of the agent variables.
Turtles will be assigned in ascending order of their who-variable.
Patches will be assigned in lines from upper left to lower right.
Since the arguments of this primitive are repeatable, don’t forget the parentheses around the statement.
;; creates an R-list "agentlist1" with the size and the id of turtles, don't forget the parentheses
(r:putagent "agentlist1" turtles "size" "who")
show r:get "agentlist1$who"
;; creates an R-list "agentlist2" with the pcolor, pxcor and pycor of patches
(r:putagent "agentlist2" patches "pcolor" "pxcor" "pycor")
show r:get "agentlist2$pcolor"
Same as r:putagent but creates an R data.frame instead a list.
Please read the notes about data.frames if one of your agent variables contains NetLogo Lists.
;; creates an R-list "agentlist2" with the pcolor, pxcor and pycor of patches, don't forget the parentheses
(r:putagentdf "df1" patches "pcolor" "pxcor" "pycor")
show r:get "class(df1)"
Same as r:putnamedlist but creates an R data.frame instead of a list.
If you send more than one list to NetLogo and the lists are of different length, the smaller ones will be filled with NA values.
If you send nested LogoLists (e.g. of type: [ [ ] [ ] … ]) to one column please read the notes about data frames with vectors in cells.
;; creates an R-list "agentlist2" with the pcolor, pxcor and pycor of patches, don't forget the parentheses
(r:putdataframe "df1" "v1" [12 13 14 15 16] "v2" ["foo1" "foo2" "foo3" "foo4" "foo5"] "v3" [1.1 2.2 3.3 4.4 5.5])
show r:get "df1$v3"
Creates a new list in R with the name name. Variable is repeatable and can be a Number, Boolean or List. Each “Variable” will get the name of its position (1, 2, 3,…). Since the arguments of this primitive are repeatable, don’t forget the parentheses around the statement.
;; creates an R-list "list1", don't forget the parentheses
(r:putlist "list1" 25.5 [25 43 32 53] "testvalue" [44.3 32.32 321.2 4.2])
show r:get "class(list1)"
show r:get "list1[[1]]"
show r:get "list1$'0'"
show r:get "list1[[2]]"
Creates a new named list in R with the name name.
Variable names and values follow in alternating sequence and may be repeated as many times as desired.
Values can be a Number, Boolean or List.
Each value will get the name varname.
Since the arguments of this primitive are repeatable, don’t forget to put the statement into parentheses.
;; creates an R-list "list1" , don't forget the parentheses
(r:putnamedlist "list1" "v1" 25.5 "v2" [25 43 32 53] "v3" "testvalue" "v4" [44.3 32.32 321.2 4.2])
show r:get "class(list1)"
show r:get "list1[[1]]"
show r:get "list1$v1"
To open an R plot in a window you can use the JavaGD plot device. With this primitive you can activate this device and all following calls of R plots will be printed with this device.
To use this device, you have to install the JavaGD package in R. Open an R terminal or the
InteractiveShell (see r:interactiveShell) and type install.packages("JavaGD").
With this plot window you can save the plot to an file of different graphic type and you can copy the plot to the clipboard. Please note, that on Linux OS it can be necessary to allow to add images to the clipboard (e.g. in KDE you have to configure KLIPPER to allow images). The resolution for raster images depends on the size of the plot window. If you need high resolution maximaze the window (and don’t use jpeg, because the driver is bad) or better use a vector image format.
Please see the notes about plotting for other details.
;; activate the JavaGD plot device
r:setPlotDevice
Stops the R engine. This is needed (only) if NetLogo is running in headless mode, for example when running BehaviorSpace experiments from the command line with something like this:
java -cp NetLogo.jar org.nlogo.headless.Main --model mymodel.nlogo --experiment exp1 --table outtab1.csv
Should be the last call in headless simulation. See usage notes above for details.
r:stop
Below are some common problems and some ideas on how to remedy them. Please keep in mind that we plan to continue to improve the R extension following the release of NetLogo 6. We welcome feedback on how to improve the extension as well as bug reports pointing us to any new problems you encounter.
See, for example, this post.
setwd()) NetLogo doesn’t find the extensionChanging working directory in R doesn’t work because it changes also Java’s library path that NetLogo needs to find its extensions. Please use absolute path to any files in R instead of changing the working directory.
R_HOME environment variable or a properties file, but couldn’t be found at the specified path. See above for how to specify R home.user.properties file, ensure that jri.home.paths includes the path given by R when you run system.file("jri",package="rJava")R_HOME. Check your user.properties file to ensure that r.home specifies a valid path to the R extension. You may also be able to work around this error by setting the R_HOME environment variable. If this error persists, please report it!Thiele, JC; Grimm, V (2010). NetLogo meets R: Linking agent-based models with a toolbox for their analysis. Environmental Modelling and Software, Volume 25, Issue 8: 972 - 974 [DOI: 10.1016/j.envsoft.2010.02.008]
The R extension is Copyright (C) 2009-2016 Jan C. Thiele and Copyright (C) 2016 Uri Wilensky / The Center for Connected Learning.
NetLogo-R-Extension is free software; you can redistribute it and/or modify it under the terms of the GNU General Public License as published by the Free Software Foundation; either version 2 of the License, or (at your option) any later version.
This program is distributed in the hope that it will be useful, but WITHOUT ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the GNU General Public License for more details.
You should have received a copy of the GNU General Public License along with NetLogo-R-Extension (located in GPL.txt). If not, see http://www.gnu.org/licenses/.